
Publications
A full list of publications can be found on Google Scholar

Chemical interactions in polyethylene glycol-induced condensates lead to an anomalous FRET response from a flexible linker-fluorescent protein crowding sensor
DOI: 10.64898/2026.02.16.706251
PMID:
[34] Mohapatra, A.; Antarasen, J.; Latham, D.R.; Barilla, M.A.; Davis, C.M.; Kisley, L. “Chemical interactions in polyethylene glycol-induced condensates lead to an anomalous FRET response from a flexible linker-fluorescent protein crowding sensor” bioRxiv.
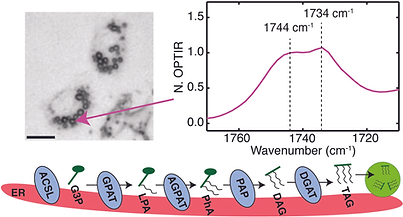
Optical photothermal infrared imaging of fatty acid metabolism in the ER of living cells
DOI: 10.64898/2025.12.09.693044
PMID:
[33] Castillo, H.B.; Davis, C.M. “Optical photothermal infrared imaging of fatty acid metabolism in the ER of living cells” bioRxiv.

Phase separation oppositely modulates G-quadruplex and i-motif DNA folding in the nuclei of living cells
DOI: 10.1101/2025.06.30.661982
PMID:
[32] Patel, B.; Hoang, C.; Yoo, H.; Davis, C.M. “Phase separation oppositely modulates G-quadruplex and i-motif DNA folding in the nuclei of living cells” bioRxiv.

Conformation and sequence determinants in the lipid binding of an adhesive peptide derived from Vibrio cholerae biofilm
DOI: 10.1101/2025.07.14.664771
PMID:
[31] Huang, X.; Prasad, R.; Saluja, S.; Yang, Y.; Yan, Q.; Shuster, S.O.; Olson, R.; Lin, C.; Davis, C.M.; Jiang, X.; Zhou, H.-X.; Yan, J. “Conformation and sequence determinants in the lipid binding of an adhesive peptide derived from Vibrio cholerae biofilm” PLoS Pathog. 2026 in press

Photopatternable thermochromism enabled by an anthracene heterodimer ligand
DOI: 10.1021/jacs.5c22789
PMID: 41687067
[30] Schreiber, E.; Logelin, M.E.; Zhang, C.; Chen, D.; Wat, J.H.; Lam, E.S.; Popofsky, M.Y.; Guo, P.; Zhu, T.; Davis, C.M.; Bartholomew, A.K. “Photopatternable thermochromism enabled by an anthracene heterodimer ligand” J. Am. Chem. Soc. 2026 in press

Weak, specific chemical interactions dictate barnase stability in diverse cellular environments
DOI: 10.1002/pro.70128
PMID: 40248880
[29] Tahir, U.; Davis, C.M. “Weak, specific chemical interactions dictate barnase stability in diverse cellular environments,” Protein Sci. 2025 34 (5) e70128.

Optical photothermal infrared imaging using metabolic probes in biological systems
DOI: 10.1021/acs.analchem.4c03752
PMID: 40207400
[28] Shuster, S.O.; Curtis, A.E.; Davis, C.M. “Optical photothermal infrared imaging using metabolic probes in biological systems,” Anal. Chem. 2025, 97 (15): 8202-8212.

Identifying the minimal sets of distance restraints for FRET-assisted protein structural modeling
DOI: 10.1002/pro.5219
PMID: 39548730
[27] Liu, Z.; Grigas, A.T.; Sumner, J.; Knab, E.; Davis, C.M., O'Hern, C.S. “Identifying the minimal sets of distance restraints for FRET-assisted protein structural modeling,” Protein Sci. 2024, 33 (12) e5219.

Similarity metrics for subcellular analysis of FRET microscopy videos
PMID: 39186078
[26] Burke, M.; Batista, V.*; Davis, C.M.* “Similarity metrics for subcellular analysis of FRET microscopy videos,” J. Phys. Chem. B. 2024, 128 (35) 8344-8354.

Exfoliation of a metal–organic framework enabled by post-synthetic cleavage of a dipyridyl dianthracene ligand
DOI: 10.1039/D4SC03524K
PMID: 39246333
[25] Logelin, M.E.; Schreiber, E.; Mercado, B.Q.; Burke, M.J.; Davis, C.M.; Bartholomew, A.K. “Exfoliation of a metal-organic framework enabled by post-synthetic cleavage of a dipyridyl dianthracene ligand,” Chem. Sci. 2024, 15 (37) 15198-15204.

Oleic acid differentially affects lipid droplet storage of de novo synthesized lipids in hepatocytes and adipocytes
DOI: 10.1039/D3CC04829B
PMID: 37873279
[24] Castillo, H.B. †; Shuster, S.O. †; Tarekegn, L.H.; Davis, C.M. “Oleic acid differentially affects lipid droplet storage of de novo synthesized lipids in hepatocytes and adipocytes,” Chem. Comm. 2024, 60: 3138-3141.

Site-specific crosslinking reveals phosphofructokinase-L inhibition drives self-assembly and attenuation of protein interactions
DOI: 10.1016/j.jbior.2023.100987
PMID: 38706136
[23] Sivadas, A.†; McDonald, E.F.†; Shuster, S.O.; Davis, C.M.; Plate, L. “Site-specific crosslinking reveals phosphofructokinase-L inhibition drives self-assembly and attenuation of protein interactions,” Adv. Biol. Regul. 2023, 90: 100987.

Chemical interactions modulate λ6-85 stability in cells
DOI: 10.1002/pro.4698
PMID: 37313657
[22] Knab, E.; Davis, C.M. “Chemical interactions modulate λ6-85 stability in cells,” Protein Sci. 2023, 32 (7): e4698.

Spatiotemporal heterogeneity of de novo lipogenesis in fixed and living single cells
PMID: 36976708
[21] Shuster, S.O.; Burke, M.J.; Davis, C.M. “Spatiotemporal heterogeneity of de novo lipogenesis in fixed and living single cells,” J. Phys. Chem. B, 2023, 127 (13): 2918-2926.

An in vitro cytomimetic of in-cell RNA folding
PMID: 35999178
[20] Yoo, H.; Davis, C.M. “An in vitro cytomimetic of in-cell RNA folding,” ChemBioChem. 2022, 23 (20): e202200406.

Spliceosomal SL1 RNA binding to U1-70k: the role of the extended RRM
DOI: 10.1093/nar/gkac599
PMID: 35876068
[19] Gopan, G.†; Ghaemi, Z.†; Davis, C.M.; Gruebele, M. “Spliceosomal SL1 RNA binding to U1-70K: the role of the extended RRM,” Nucl. Acids Res. 2022, 50 (14): 8193-8206.

Cellular sticking can strongly reduce complex binding by speeding dissociation
PMID: 33826329
[18] Davis, C.M.*; Gruebele, M.* “Cellular sticking can strongly reduce complex binding by speeding dissociation,” J. Phys. Chem. B 2021, 125 (15): 3815-3823.

Cytoskeletal drugs modulate off-target protein folding landscapes inside cells
DOI: 10.1021/acs.biochem.0c00299
PMID: 32567840
[17] Davis, C.M.*; Gruebele, M.* “Cytoskeletal drugs modulate off-target protein folding landscapes inside cells,” Biochemistry 2020, 59 (28), 2650-2659.

An in vitro mimic of in-cell solvation for protein folding studies
DOI: 10.1002/pro.3833
PMID: 31994240
[16] Davis, C.M.*; Deutsch, J.C.; Gruebele, M.* “An in vitro mimic of in-cell solvation for protein folding studies,” Protein Sci. 2020, 29 (4), 1046-1054.
Quantifying protein dynamics and stability in a living organism
DOI: 10.1038/s41467-019-09088-y
PMID: 30862837
[15] Feng, R.; Gruebele, M.*; Davis, C.M.*“Quantifying protein dynamics and stability in a living organism,” Nat. Commun. 2019, 10, 1179.

Cell volume controls protein stability and compactness of the unfolded state
PMID: 30289261
[14] Wang, Y.†; Sukenik, S.*†; Davis, C.M.; Gruebele, M.* “Cell volume controls protein stability and compactness of the unfolded state,” J. Phys. Chem. B 2018, 122 (49), 11762-11770.

A quantitative connection of experimental and simulated folding landscapes by vibrational spectroscopy
DOI: 10.1039/C8SC03786H
PMID: 30647892
[13] Davis, C.M.†; Polzi, L.Z.†; Gruebele, M.; Amadei, A.; Dyer, R.B.*; Daidone, I.* “A quantitative connection of experimental and simulated folding landscapes by vibrational spectroscopy,” Chem. Sci. 2018, 9, 9002-9011.

Soluble zwitterionic poly(sulfobetaine) destabilizes proteins
DOI: 10.1021/acs.biomac.8b01120
PMID: 30064224
[12] Kisley, L.; Serrano, K.M.; Davis, C.M.; Guin, D.; Murphy, E.; Gruebele, M.*; Leckband, D.E.* “Soluble zwitterionic poly(sulfobetaine) destabilizes proteins,” Biomacromolecules 2018, 19 (9), 3894-3901.

Non-steric interactions predict the trend and steric interactions the offset of protein stability in cells
PMID: 29877016
[11] Davis, C.M.; Gruebele, M. “Non-steric interactions predict the trend and steric interactions the offset of protein stability in cells,” ChemPhysChem 2018, 19 (18), 2290-2294.

Labeling for quantitative comparison of imaging measurements in vitro and in cells
DOI: 10.1021/acs.biochem.8b00141
PMID: 29546761
[10] Davis, C.M.*; Gruebele, M.* “Labeling for quantitative comparison of imaging measurements in vitro and in cells,” Biochemistry 2018, 57 (13), 1929-1938.

Binding, folding, and insertion of a β-hairpin peptide at a lipid bilayer surface: influence of electrostatics and lipid tail packing
DOI: 10.1016/j.bbamem.2017.12.019
PMID: 29291379
[9] Reid, K.; Davis, C.M.; Dyer, R.B.; Kindt, J.T. “Binding, folding, and insertion of a β-hairpin peptide at a lipid bilayer surface: influence of electrostatics and lipid tail packing,” Biochim. Biophys. Acta Biomembr. 2018, 1860 (3), 792-800.

How does solvation in the cell affect protein folding and binding?
DOI: 10.1016/j.sbi.2017.09.003
PMID: 29035742
[8] Davis, C.M.; Gruebele, M.; Sukenik, S. “How does solvation in the cell affect protein folding and binding?” Curr. Opin. Struct. Biol. 2018, 48, 23-29.

Parallel folding pathways of Fip35 WW domain explained by infrared spectra and their computer simulation
PMID: 28881468
[7] Polzi, L.Z.; Davis, C.M.; Gruebele, M.; Dyer, R.B.; Amadei, A.; Daidone, I. “Parallel folding pathways of Fip35 WW domain explained by infrared spectra and their computer simulation,” FEBS Lett. 2017, 591 (20), 3265-3275.

Dual time-resolved temperature-jump fluorescence and infrared spectroscopy for the study of fast protein dynamics
DOI: 10.1016/j.saa.2017.01.069
PMID: 28189834
[6] Davis, C.M.; Reddish, M.J.; Dyer, R.B. “Dual time-resolved temperature-jump fluorescence and infrared spectroscopy for the study of fast protein dynamics,” Spectrochim. Acta A 2017, 178, 185-191.
The role of electrostatic interactions in folding of β-proteins
DOI: 10.1021/jacs.5b13201
PMID: 26750867
[5] Davis, C.M.; Dyer, R.B. “The role of electrostatic interactions in folding of β-proteins,” J. Am. Chem. Soc. 2016, 138 (4), 1456-1464.
Fast helix formation in the B domain of protein A revealed by site-specific infrared probes
DOI: 10.1021/acs.biochem.5b00037
PMID: 25706439
[4] Davis, C.M.; Cooper, A.K.; Dyer, R.B. “Fast helix formation in the B domain of protein A revealed by site-specific infrared probes,” Biochemistry 2015, 54 (9), 1758-1766.
WW Domain folding complexity revealed by infrared spectroscopy
DOI: 10.1021/bi500556h
PMID: 25121968
[3] Davis, C.M.; Dyer, R. B. “WW Domain folding complexity revealed by infrared spectroscopy,” Biochemistry 2014, 53 (34), 5476-5484.
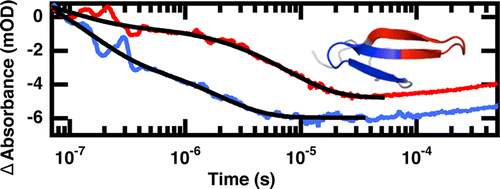
Dynamics of an ultrafast folding subdomain in the context of a larger protein fold
DOI: 10.1021/ja409608r
PMID: 24320936
[2] Davis, C.M.; Dyer, R. B. “Dynamics of an ultrafast folding subdomain in the context of a larger protein fold,” J. Am. Chem. Soc. 2013, 135 (51), 19260-19267.
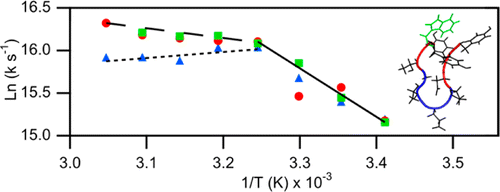
Raising the speed limit for β-hairpin formation
DOI: 10.1021/ja3046734
PMID: 22873643
[1] Davis, C.M.; Xiao, S.; Raleigh, D.P.*; Dyer, R. B.* “Raising the speed limit for β-hairpin formation,” J. Am. Chem. Soc. 2012, 134 (35), 14476-14482.


